Nusara Yinyom
Department of Microbiology, Faculty of Medical Science, Naresuan University, Phitsanulok, Thailand
Kusuma Rintachai
Department of Civil Engineering, Faculty of Engineering, Naresuan University, Phitsanulok, Thailand
Siriwan Wichai
Department of Microbiology, Faculty of Medical Science, Naresuan University, Phitsanulok, Thailand
Keywords: methanogenesis archaea biogenic gas methanogenic media phylogenetic analysis MPN
Abstract
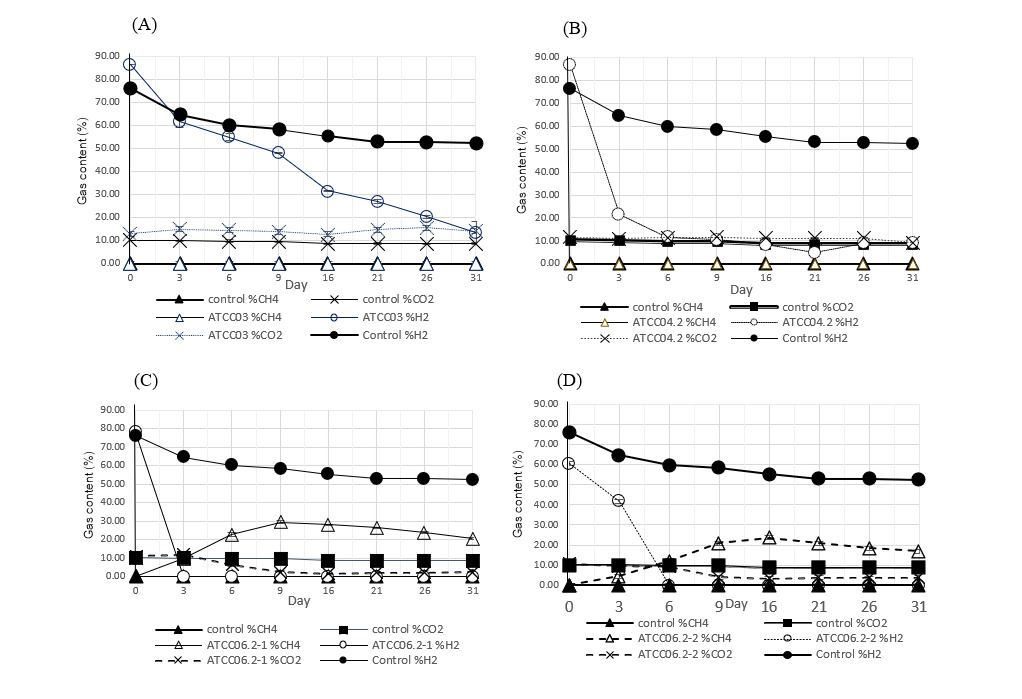
Some depleted oil and gas fields harbor anaerobic microbes, including methanogens. In this study, methanogens were isolated from a depleted oil and gas field in a neighboring country using enrichment techniques in methanogenic media (ATCC 1340). Five isolates were obtained. Among them, three isolates- ATCC 06.2-1, ATCC 06.2-2, and ATCC reactor 1- demonstrated a strong ability to produce high levels of methane in the ATCC 1340 medium. This culture medium is specifically designed to convert H2 and CO2 at a ratio of 4:1, leading to a theoretical total yield of 249.82 µmol of methane. The results indicated that ATCC 06.2-1, ATCC 06.2-2, and ATCC reactor 1 achieved methane production percentages of 94.07%, 75.65%, and 72.05% of the theoretical yield, respectively. Additionally, all three isolates could utilize formate and methanol as substrates for methane production and were classified as hydrogenotrophs. Genetic identification revealed that the three methane-producing isolates- ATCC 06.2-1, ATCC 06.2-2, and ATCC reactor 1- belong to the species Methanobacterium ferruginis Mic6c05T. The other two isolates, ATCC 03 and ATCC 04.2, were identified as Clostridium sporogenes DSM 795T. The three methane-producing isolates and the two Clostridium isolates show significant potential for optimizing alternative energy sources.
How to Cite
Yinyom, N., Rintachai, K., & Wichai, S. (2025). High methane production by Methanobacterium ferruginis Mic6c05T, a hydrogenotrophic methanogen isolated from a depleted oil and gas field in a neighboring country. Asia-Pacific Journal of Science and Technology, 30(06), APST–30. https://doi.org/10.14456/apst.2025.93
References
Mei R, Kaneko M, Imachi H, Nobu MK. The origin and evolution of methanogenesis and archaea are intertwined. PNAS Nexus. 2023;2(2):1-10.
Wang J, Yao X, Xu H, Lou H, Hu B. Methane cycle in subsurface environment: A review of microbial processes. Environ Res. 2025;265:120404.
Woraruthai T, Kunno J, Pongsopon M, Yansakon K, Phoopraintra P, Chantiwas R, et al. Identification and cultivation of hydrogenotrophic methanogens from palm oil mill effluent for high methane production. Int J Energy Res. 2020;44(13):10058–10070.
Jain KA, Suryawanshi PC, Chaudhari AB. Hydrogenotrophic methanogen strain of Methanospirillum from anaerobic digester fed with agro-industrial waste. Biologia. 2021;76:255–266.
Ray S, Kuppam C, Pandit S, et al. Biogas upgrading by hydrogenotrophic methanogens: An overview. Waste Biomass Valor. 2023;14:537–552.
Qian L, Yu X, Gu H, et al. Vertically stratified methane, nitrogen and sulphur cycling and coupling mechanisms in mangrove sediment microbiomes. Microbiome. 2023;11:70-71.
Liu Z, Mao X, Wu Y, Xia L, Yu H, Tang W, et al. Methanogenic community characteristics and its influencing factors in reservoir sediments on the northeastern qinghai plateau. Biology (Basel). 2024;13(8):61-65.
Tan J, Lichtfouse E, Luo M, Liu Y, Tan F, Zhang C, et al. Aquaculture drastically increases methane production by favoring acetoclastic rather than hydrogenotrophic methanogenesis in shrimp pond sediments. Aquaculture. 2023;563(2):738999.
Akimbekov, N. S., Digel, I., Tastambek, K. T., Kozhahmetova, M., Sherelkhan, D. K., & Tauanov, Z. Hydrogenotrophic methanogenesis in coal-bearing environments: Methane production, carbon sequestration, and hydrogen availability. Int J Hydrogen Energy. 2024;52(D):1264–1277.
Myint L. Biogenic gas potential of Myanmar. In: 4th AAPG/EAGE/MGS Myanmar oil and gas conference: Myanmar: A global oil and gas hotspot. AAPG Asia Pacific Region; 2019.
Rintachai K, Phenrat T, Wichai S, Limmongkon A, Yinyom N. Design and commissioning of continuously stirred anaerobic bioreactor for upcycling carbon dioxide (CO2) to methane (CH4) via methanogenesis. Naresuan Univ Eng J. 2023;18(2):41–50.
ATCC. ATCC Medium 1340 (MS medium for methanogens). Manassas (VA): American Type Culture Collection; 2025.
Zhou J, Smith JA, Li M, Holmes DE. Methane production by Methanothrix thermoacetophila via direct interspecies electron transfer with Geobacter metallireducens. mBio. 2023;14:e00360.
Machado RAR, Muller A, Hiltmann A, Bhat AH, Půža V, Malan AP, et al. Genome-wide analyses provide insights into genetic variation, phylo- and co-phylogenetic relationships, and biogeography of the entomopathogenic nematode genus Heterorhabditis. Mol Phylogenet Evol. 2025;204:108284.
Santaweesuk A, Artnaseaw A, Benjapiyaporn C. Optimization of methane production through co-digestion of pig manure with napier grass. Clean Eng Technol. 2025;26:100931.
Nguyen TH, et al. Using metagenomics tool to evaluate the enrichment efficiency of methanogens in marine sediment in Truong Sa archipelago, Khanh Hoa province, Vietnam. E3S Web Conf. 2023;407:2010.
Rintachai K. Feasibility study of methane production from carbon source by deep underground microorganisms in the gulf of martaban, Myanmar [Master’s thesis]. Naresuan University; 2021.
Shuai Y, Zhang S, Grasby SE, Hou W, Chen Z, Huang L, et al. Microbial consortia controlling biogenic gas formation in the Qaidam Basin of western China. J Geophys Res Biogeosci. 2016;121(8):2296–2306.
Wellinger A, Murphy J, Baxter D. The biogas handbook: Science, production and applications. Woodhead Publishing; 2013.
Hungate R. Chapter IV A roll tube method for cultivation of strict anaerobes. In: Norris JR, ribbons DW, editors. J Microbiol Methods. 1969;3:117–132.
ATCC. ATCC Medium 1045 (Methanobacteria Medium) [Internet]. Manassas (VA): American Type Culture Collection; 2025.
Karekar S, Stefanini R, Ahring B. Homo-acetogens: Their metabolism and competitive relationship with hydrogenotrophic methanogens. Microorganisms. 2022;10(2):397-399.
Susilawati R, Esterle JS, Golding SD, Mares TE. Microbial methane potential for the South Sumatra Basin coal: Formation water screening and coal substrate bioavailability. Energy Procedia. 2014;65:282–291.
Garcia-Robledo E, Ottosen LD, Voigt NV, Kofoed M, Revsbech NP. Micro-scale H2–CO2 dynamics in a hydrogenotrophic methanogenic membrane reactor. Front Microbiol. 2016;7:1276.
Wei S, Cui H, Zhang Y, Su X, Dong H, Chen F, et al. Comparative evaluation of three archaeal primer pairs for exploring archaeal communities in deep-sea sediments and permafrost soils. Extremophiles. 2019;23(6):747–757.
Brown JL, Tran-Dinh N, Chapman B. Clostridium sporogenes PA 3679 and its uses in the derivation of thermal processing schedules for low-acid shelf-stable foods and as a research model for proteolytic Clostridium botulinum. J Food Prot. 2012;75(4):779–792.
Cai G, Jin B, Monis P, Saint C. A genetic and metabolic approach to redirection of biochemical pathways of Clostridium butyricum for enhancing hydrogen production. Biotechnol Bioeng. 2013;110(1):338-342.
Gupta S, Fernandes A, Lopes A, Grasa L, Salafranca J. Microbes and parameter influencing dark fermentation for hydrogen production. Appl Sci. 2024;14(23):10789.
Albuquerque MM, Sartor GB, Martines-Burgos WJ, Scapini T, Edwiges T, Soccol CR, Medeiros ABP. Biohydrogen produced via dark fermentation: A review. Methane. 2024;3(3):500-532.
Sun X, Atiyeh HK, Hunhnke RL, Tanner RS. Syngas fermentation process development for production of biofuels and chemicals: A review. Bioresour Technol Rep. 2019;7:100279.
Christman GD, León-Zayas RI, Zhao R, et al. Novel clostridial lineages recovered from metagenomes of a hot oil reservoir. Sci Rep. 2020;10:8048.

Published: Nov 4, 2025
License
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
