Md. Habibur Rahman
Department of Fisheries Biology and Aquatic Environment, Gazipur Agricultural University, Gazipur 1706, Bangladesh
Shahittya Mitra Pranto
Department of Fisheries Biology and Aquatic Environment, Gazipur Agricultural University, Gazipur 1706, Bangladesh
Mousumi Das
Department of Aquaculture, Gazipur Agricultural University, Gazipur 1706, Bangladesh
Koushik Chakroborty
Department of Aquatic Environment and Resource Management, Sher-e-Bangla Agricultural University, Dhaka 1207, Bangladesh
Mauching Marma
Department of Fisheries Biology and Aquatic Environment, Gazipur Agricultural University, Gazipur 1706, Bangladesh
Md. Shahanoor Alam
Department of Genetics and Fish Breeding, Gazipur Agricultural University, Gazipur 1706, Bangladesh
S. M. Rafiquzzaman
Department of Fisheries Biology and Aquatic Environment, Gazipur Agricultural University, Gazipur 1706, Bangladesh
Keywords: Proteolytic, Bacillus subtilis, Enzyme, Microorganism
Abstract
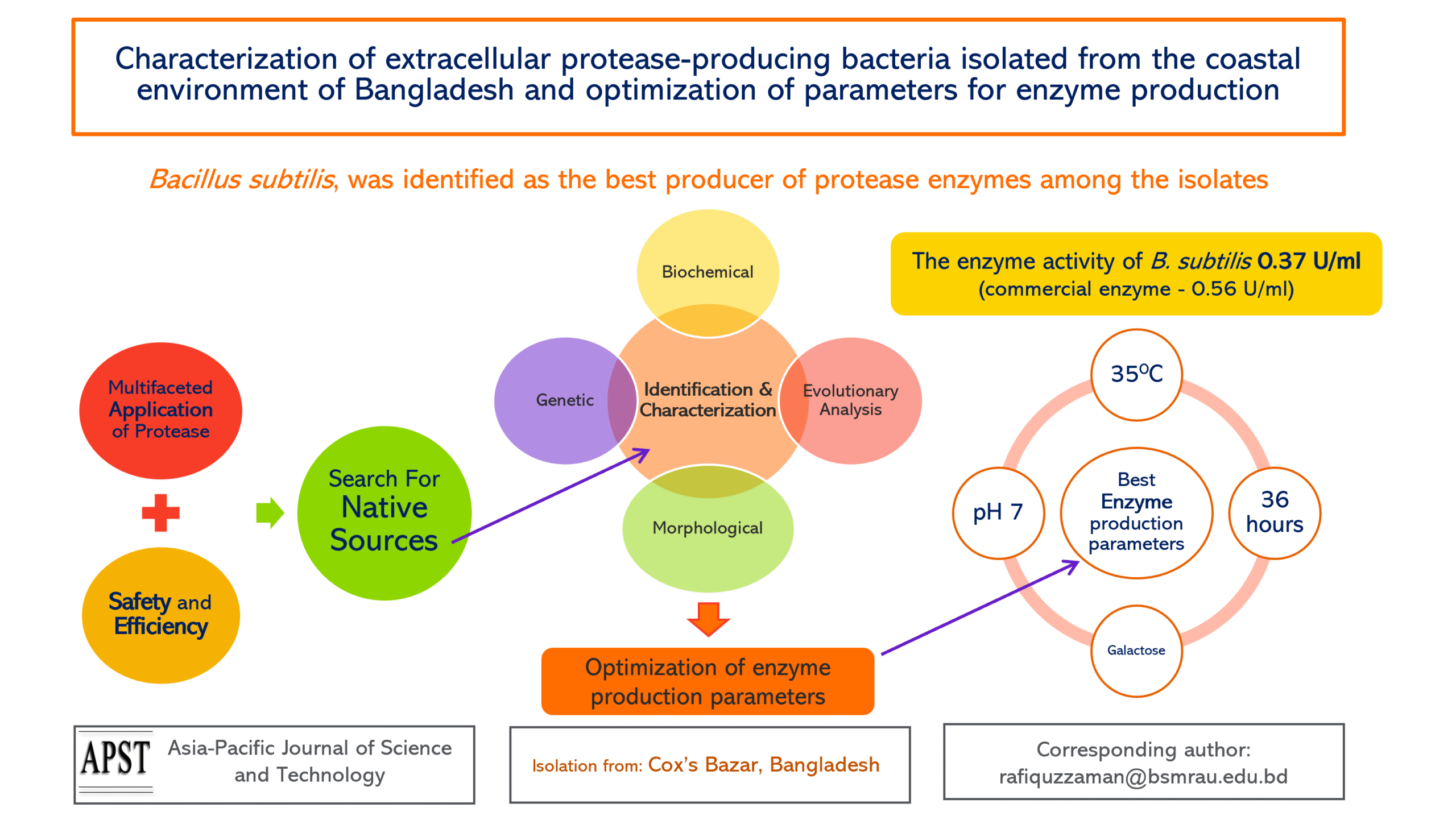
Bangladesh’s coastal soil and water harbor a diverse array of native microorganisms that can serve as a significant source of natural protease enzymes, with broad applications in biotechnology, food industries, and environmental sectors. The objective of this study was to isolate and identify native protease-producing bacteria from Cox’s Bazar, a coastal region of Bangladesh, and to optimize the parameters for enzyme production. The bacteria found in the soil and water samples were initially assessed using the zone of inhibition technique and hemolytic test. Five of the isolated bacteria exhibited proteolytic activity and were found to be non-pathogenic. The isolates were subsequently identified through morphological, biochemical, molecular, and evolutionary analysis. Among the identified isolates (Bacillus subtilis, Xenorhabdus nematophilia, and Enterobacter cloacae), B. subtilis was determined to be the most productive bacterial species. A temperature of 35°C, pH 7, 36 h of incubation, and galactose as the carbon source, were the best conditions for protease production. The enzyme activity of B. subtilis was found to be 0.37 U/mL, compared to 0.56 U/mL for a commercial enzyme. These findings suggest that B. subtilis obtained from the coastal water of Bangladesh has the potential to serve as a local source for protease production, with applications in food processing, animal feed, dairy, meat, textiles, waste management, and pharmaceutical industries in Bangladesh.
References
Razzaq A, Shamsi S, Ali A, Ali Qurban and Sajjad M, Malik A, Ashraf M. Microbial proteases applications. Front Bioeng Biotechnol 2019;7:1-10.
Badhe P, Joshi M, Adivarekar R. Optimized production of extracellular proteases by Bacillus subtilis from degraded abattoir waste. J Biosci Biotechnol. 2016;5:29–36.
Tavano OL. Protein hydrolysis using proteases: An important tool for food biotechnology. J Mol Catal B Enzym 2013;90:1–11.
Fulzele R, DeSa E, Yadav A, Shouche Y, Bhadekar R. Characterization of novel extracellular protease produced by marine bacterial isolate from the Indian Ocean. Braz J Microbiol. 2011;42:1364–73.
Nisha S, Divakaran. Optimization of alkaline protease production from Bacillus subtilis NS isolated from sea water. Afr J Biotechnol. 2014;13:1707–1713.
Bhaskar N, Sudeepa ES, Rashmi HN, Tamil Selvi A. Partial purification and characterization of protease of Bacillus proteolyticus CFR3001 isolated from fish processing waste and its antibacterial activities. Bioresour Technol 2007;98:2758–2764.
Singh P, Rani A, Chaudhary N. Isolation and characterization of protease producing Bacillus sp from soil. Int J Pharm Sci Res. 2015;6:633–639.
[8] Hossain I, Mitu IJ, Hasan MR. Industrial enzyme production in bangladesh: current landscape, scope, and challenges. Asian J Med Biol Res. 2023;9:693-695.
Wiltschi B, Cernava T, Dennig A, Galindo Casas M, Geier M, Gruber Steffen and Haberbauer M, et al. Enzymes revolutionize the bioproduction of value-added compounds: From enzyme discovery to special applications. Biotechnol Adv. 2020;40:107520.
Harley JP, Prescott LM. Harley Prescott: 7th edition. Laboratory Exercises in Microbiology. 2002.
Singhal N, Kumar M, Kanaujia PK, Virdi JS. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol. 2015;6:144398.
Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526.
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chem.1951;193:265–275.
Alnahdi HS. Isolation and screening of extracellular proteases produced by new Isolated Bacillus sp. J Appl Pharm Sci. 2012;1:2-71.
Das G, Prasad MP. Isolation, purification & mass production of protease enzyme from Bacillus subtilis. Intl Res J Microbiol. 2010;1:26–31.
[Mahajan PM, Nayak S, Lele SS. Fibrinolytic enzyme from newly isolated marine bacterium Bacillus subtilis ICTF-1: Media optimization, purification and characterization. J Biosci Bioeng. 2012; 113:307–714.
Hariharan A, Rajadurai UM, Palanivel I. Isolation, purification and mass production of protease from Bacillus subtilis. SSRN Electron J. 2019;10:21-39.
Uddin ME, Rahman M, Faruquee HM, Khan MRI, Ahammed T, Alam MF, et al. Identification, partial purification and characterization of protease enzyme from locally isolated soil bacterium. Am Res J Bio Sci. 2015;1(3):1-9.
Padmapriya M, Williams BC. Purification and characterization of neutral protease enzyme from Bacillus subtilis. J Microbiol Biotechnol Res. 2012;2:612–628.
Sen S, Dasu VV, Dutta K, Mandal B. Characterization of a novel surfactant and organic solvent stable high-alkaline protease from new Bacillus pseudofirmus SVB1. Res J Microbiol 2011;6:76-79.
Palsaniya P, Mishra R, Beejawat N, Sethi S, Gupta BL. Optimization of alkaline protease production from bacteria isolated from soil. J Microbiol Biotechnol Res 2012;2:858–865.
Liu Z, Liu G, Guo X, Li Y, Ji N, Xu X, et al. Diversity of the protease-producing bacteria and their extracellular protease in the coastal mudflat of Jiaozhou Bay, China: in response to calm naturally growing and aquaculture. Front Microbiol 2023;14:1164937.
Dreyer J, Malan AP, Dicks LMT. Bacteria of the genus Xenorhabdus, a novel source of bioactive compounds. Front Microbiol 2018;9:3177.
Fajingbesi AO, Anzaku AA, Akande M, Emmanuel I A and Akwashiki O. Production of protease enzyme from fish guts using Pseudomonas fluorescens, Enterobacter cloacae and Bacillus megaterium. J Clin Pathol Lab Med. 2018;2:1–7.
Rupali D. Screening and isolation of protease producing bacteria from soil collected from different areas of Burhanpur Region (MP) India. Int J Curr Microbiol Appl Sci. 2015;4:597–606.
Sahin S, Demir Y, Ozmen I. Production of protease from Bacillus subtilis under SSF and effect of organic solvents on lyophilized protease preparations. Int J Chem Res. 2020;1:1–6.
Patel AR, Mokashe NU, Chaudhari DS, Jadhav AG, Patil UK. Production optimization and characterization of extracellular protease secreted by newly isolated Bacillus subtilis AU-2 strain obtained from Tribolium castaneum gut. Biocatal Agric Biotechnol 2019;19:1-8.
Shivanand P, Jayaraman G. Production of extracellular protease from halotolerant bacterium, Bacillus aquimaris strain VITP4 isolated from Kumta coast. Process Biochem. 2009;44:1088–1094.
Pant G, Prakash A, Pavani JVP, Bera S, Deviram GVNS, Kumar A, et al. Production, optimization and partial purification of protease from Bacillus subtilis. J Taibah Univ Sci. 2015;9:50–55.
Deviga JT, Ishwarya R, Geetha M, Sangeetha D. Studies on the optimization of protease production by thermophilic Bacillus subtilis isolated from raw milk sample. Bull Environ Pharmacol Life Sci. 2022; 12:91–98.
Gerze A, Omay D, Guvenilir Y. Partial purification and characterization of protease enzyme from Bacillus subtilis megatherium. Appl Biochem Biotechnol. 2009;121:335–345.
Choi JW, Song N-E, Hong S, Rhee YK, Hong H-D, Cho C-W. Engineering Bacillus subtilis J46 for efficient utilization of galactose through adaptive laboratory evolution. AMB Express. 2024;14:1-4.
Suganthi C, Mageswari A, Karthikeyan S, Anbalagan M and Sivakumar A, Gothandam KM. Screening and optimization of protease production from a halotolerant Bacillus licheniformis isolated from saltern sediments. J Genetic Eng Biotechnol. 2013;11:47–52.
Blázquez B, San León D, Rojas A, Tortajada M, Nogales J. New insights on metabolic features of Bacillus subtilis based on multistrain genome-scale metabolic modeling. Int J Mol Sci. 2023;24:70-91.
Demirkan E, Kut D, Sevgi T, Dogan M, Baygin E. Investigation of effects of protease enzyme produced by Bacillus subtilis 168 E6-5 and commercial enzyme on physical properties of woolen fabric. J Text Inst. 2019;111(1):26-35.

License
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
