Vijay Lobo
Department of Biochemistry, Research Development Cell (RDC), PRIST Deemed University, Thanjavur, Tamil Nadu, India.
Amjath Khan
Department of Biochemistry, Research Development Cell (RDC), PRIST Deemed University, Thanjavur, Tamil Nadu, India.
Subhashini Ramakrishnan
Department of Biotechnology, Dr.G.R.Damodaran college of Science, Coimbatore – 641014, Tamil Nadu, India.
Ramathilaga Ariyamuthu
Department of Biotechnology, Dr.G.R.Damodaran college of Science, Coimbatore – 641014, Tamil Nadu, India.
Balambigai Narayanasamy
Department of Biotechnology, Dr.G.R.Damodaran college of Science, Coimbatore – 641014, Tamil Nadu, India.
Bakrudeen Ali Ahmed Abdul
Department of Biochemistry, Research Development Cell (RDC), PRIST Deemed University, Thanjavur, Tamil Nadu, India.
DOI: https://doi.org/10.14456/apst.2025.32
Keywords: Microsporum gypseum Subtilisin-like protease1 AlphaFold Cyanidin-3-O-rhamnoside docking MD simulation
Abstract
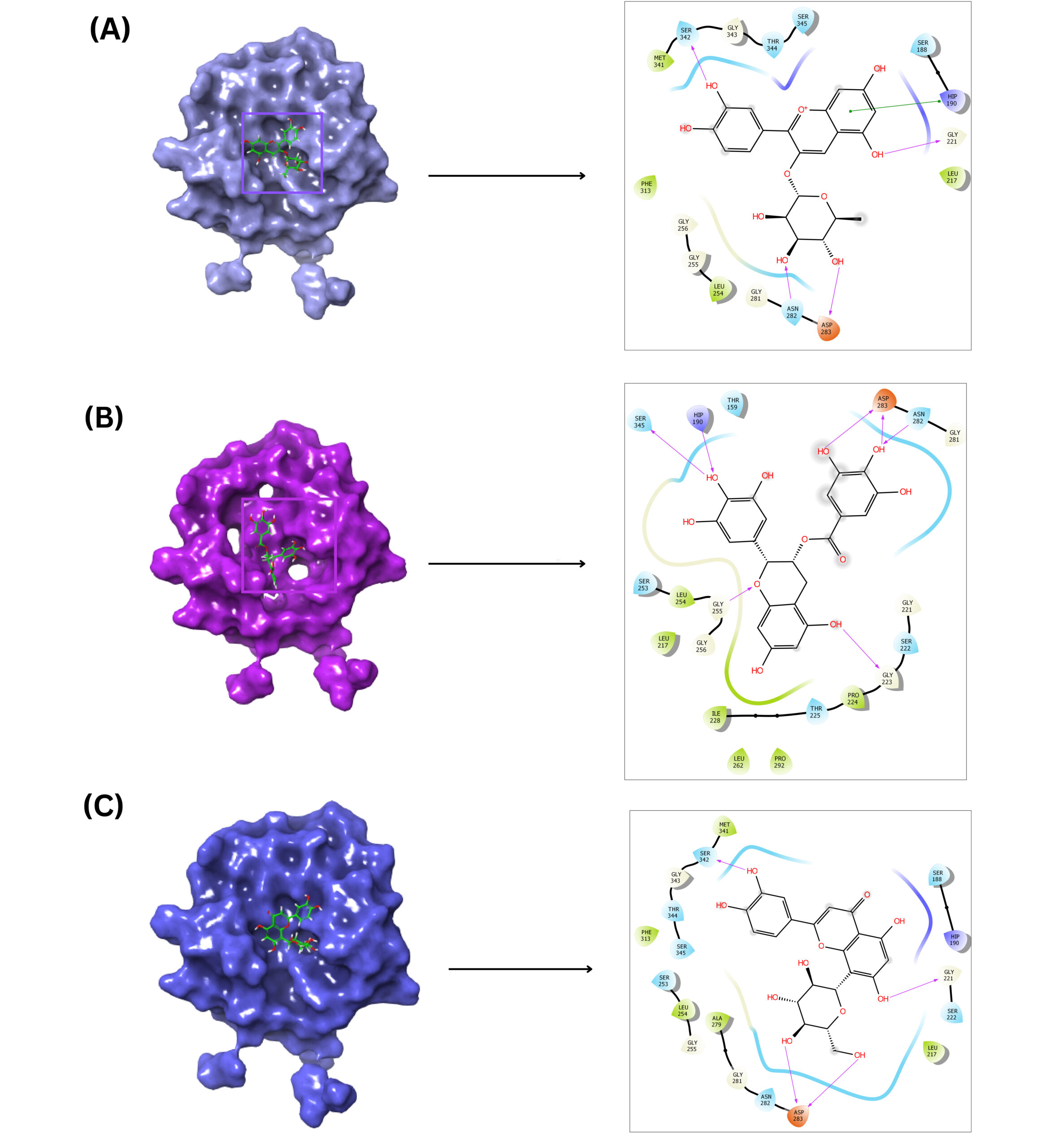
Microsporum gypseum, a keratinophilic fungus belonging to the dermatophyte group causes infections of the skin, hair, and nails in humans and animals, leading to conditions such as tinea capitis, tinea corporis, and tinea faciei. Virulence factors play a crucial role in the pathogenicity and subsequent host tissue damage. In dermatopytes, proteases are the primary virulence factors, which facilitate host invasion and utilization of the stratum corneum. Unlike most studies targeting the ergosterol biosynthesis pathway for the treatment of fungal infections, this research focuses on a major virulence enzyme, subtilisin-like protease (SUB-1), employing in silico evaluation of antifungal compounds against this enzyme. The three-dimensional structure of SUB-1 was retrieved from the AlphaFold database and evaluated using the prediction local distance difference test (pLDDT) scores and a Ramachandran plot. Active site residues were identified based on a literature review and using the webserver ConSurf. Structures of compounds were downloaded from the PubChem database, and the physicochemical properties were analyzed using the Swiss ADME database. Selected compounds were then docked with the SUB-1 by using Glide software, and molecular dynamics simulations were conducted using the Desmond module of Schrödinger for identifying the best-docked complex structure. The results revealed that cyanidin-3-O-rhamnoside exhibits potent activity against SUB-1, with a docking score of -9.4 kcal/mol, binding free energy of -55.23 kcal/mol, and interactions with the active site residues H190, N282, and H342. The efficiency of this compound in inhibiting the growth of M. gypseum needs to be validated further using experimental studies.
How to Cite
Lobo, V. ., Khan, A. ., Ramakrishnan, S. ., Ariyamuthu , R. ., Narayanasamy , B. ., & Ali Ahmed Abdul, B. . (2025). Cyanidin-3-O-rhamnoside: A promising inhibitor of the virulence protein subtilisin-like protease-1 in Microsporum gypseum. Asia-Pacific Journal of Science and Technology, 30(02), APST–30. https://doi.org/10.14456/apst.2025.32
References
Simpanya MF. Dermatophytes: Their taxonomy, ecology and pathogenicity. Rev Iberoam Micol. 2000; 17:1-12.
Ginter G.Ecology, epidemiology and clinical symptomatology of infections due Microsporum gypseum. Mycoses. 2009; 32(10): 531-535.
Hube BR, Hay J, Brasch J, Veraldi S, Schaller M. Dermatomycoses and inflammation: the adaptive balance between growth, damage, and survival. J Mycol Med.2015; 25(1): e44–58.
AkcaglarS, Ener B, Toker SC, Ediz B, Tunali S, Tore O. A comparative study of dermatophyte infections in Bursa, Turkey.Med Mycol. 2011; 49:602-607.
Monod M. Secreted proteases from dermatophytes. Mycopathologia. 2008; 166(5–6): 285–94.
Descamps F, Brouta F, Monod M, Zaugg C, Baar D, Losson B, Mignon B. Isolation of a Microsporumcanis gene family encoding three subtilisin like proteases expressed in vivo. J Invest Dermatol. 2002; 119: 830-835.
Al-shibly MK, Al-Marshedy AM. Molecular study of virulence factors influencing the pathogenicity of Trichophyton rubrum. Ecology, Environment and Conservation. 2019; 25(1): 153-158.
Li J, Li Y, Yang J, Dong L, Tian B, Yu Z, Liang L, Zhang Y, Wang X, Zhang K. New insights into the evolution of subtilisin-like serine protease genes in Pezizomycotina. BMC Evolutionary Biology. 2010; 10:1-14.
Suntar I. Importance of ethnopharmacological studies in drug discovery: role of medicinal plants. Phytochem Rev, 2020;19, 1199–1209.
Kinghorn AD. Pharmacognosy in the 21st century. J Pharm Pharmacol. 2001; 53 (2): 135 – 148.
Balakumar S, Rajan S, Thiyagarajan T, Jeeva S. Antifungal activity of Ocimum sanctum Linn. (Lamiaceae) on clinically isolated dermatophytic fungi. Asian Pac J Trop Med. 2011; 4(8): 654-7.
Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, Yuan D, Stroe O, et al. Alpha fold protein structure database: massively expanding the structural coverage ofprotein-sequence space with high-accuracy models. Nucleic Acids Res 2022; 50: D439–D444.
Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK – a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993; 26 (2):283–291.
Ashkenazy H. et al.ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016; 44(W1): W344-50.
Riyadi PH, Romadhon, Sari ID, Kurniasih RA, Agustini TW, Swastawati F, Herawati VE Tanod WA. Swiss ADME predictions of pharmacokinetics and druglikeness properties of small molecules present in Spirulina platensis. Conf. Ser.: Earth Environ. Sci., 2021; 890.
Castro-Alvarez A, Costa AM, Vilarrasa J. The performance of several docking programs at reproducing protein–macrolide-like crystal structures. Molecules. 2017; 22 (136), 1–14.
Wang E, Sun H, Wang J, Wang Z, Liu H, Zhang JZH, Hou T. End-Point Binding Free Energy Calculation with MM/PBSA and MM/GBSA: Strategies and applications in drug design. Chem Rev. 2019; 119:9478-9508.
Blessy JJ, Sharmila DJS. Molecular simulation of N-acetylneuraminic acid analogs and molecular dynamics studies of cholera toxin-Neu5Gc complex. J Biomol Struct Dyn. 2014; 33(5): 1126-1139.
Fu Y, Zhao J and Chen Z. Insights into the molecular mechanisms of protein-ligand interactions by molecular docking and molecular dynamics simulation: A case of oligopeptide binding protein. Comput Math Methods Med. 2018; 3502-14.
Peng Y, Yang Y, Li L, Jia Z, Cao W Alexov E. DFMD: Fast and effective Delphi force steered molecular dynamics approach to model ligand approach toward a receptor: Application to spermine synthase enzyme. Front Mol Biosci. 2019; 6: 1-12.
Moskaluk AE, VandeWoude S. Current topics in dermatophyte classification and clinical diagnosis. Pathogens. 2022, 11, 957.
Lee WJ, Park JH, Kim JY, Jang YH, Lee S, Bang YJ, Jun JB. Low but continuous occurrence of Microsporum gypseum infection in the study on 198 cases in South Korea from 1979 to 2016. Ann. Dermatol. 2018; 30: 427–431.
Hay, R. Therapy of Skin, Hair and Nail Fungal Infections. J Fungi. 2018. 4(99): 1-13.
Descamps F, Brouta F, Monod M, Zaugg C, Baar D, Losson B, Mignon B. Isolation of a Microsporumcanis gene family encoding three subtilisin like proteases expressed in vivo. J Investig Dermatol 2002; 119: 830-835.
Moallaei H, Zaini F, Rezaie S, Nourbakhsh F, Larcher G. The enzymatic activity and molecular characterization of a secreted subtilisin-like protease in Microsporumgy pseum and Tricho phytonvan breuseghemii. Iranian J Publ Health. 2009; 38: 25-33.
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. Highly accurate protein structure prediction with Alpha Fold. Nature.2021; 596: 584–589.
Mishra P, Wardhan V, Pandey A, Chakraborty S, Garg G, Chakraborty N. Comparative analysis of sequence-structure function relationship of the SUN-Domain Protein CaSUN1. J Phylogenetics Evol Biol. 2017; 5:3.
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 1997; 23:3-25.
Hussain MSA, Velusamy S, Muthusamy J. Balanitesaegyptiaca (L.) Del. for dermatophytose: Ascertaining the efficacy and mode of action through experimental and computational approaches. Inform Med Unlocked 2019; 15:1-11.
Abuthakir MHS, Sharmila V, Jeyam M. Screening Balanitesaegyptiacafor inhibitors against putative drug targets in Microsporum gypseum– Subtractive proteome, docking and simulation approach. Infect Genet Evol. 2021; 90(3):104755.
Al Aboody MS and Mickymaray S. Anti-fungal efficacy and mechanisms of flavonoids. Anitbiotics. 2020; 9(2): 45.
Park BJ, Taguchi H, Kamei K, Matsuzawa T, Hyon S, Park J. In Vitro Antifungal Activity of Epigallocatechin 3- O -Gallate against clinical isolates of dermatophytes. Yonsei Med J. 52(3):535-8.
De Campos MP, Filho VC, Da SilvaRZ, Yunes RA, Zacchino S, Juarez S, Cruz RCB, Cruz AB. Evaluation of antifungal Activity of Piper solmsianum C. DC. var.solmsianum (Piperaceae). Biol Pharm Bull. 2005; 28(8) 1527—1530.
Ghoreishi M, Heidari E. Extraction of epigallocatechin gallate from green tea via modified supercritical CO2: Experimental, modeling and optimization. J Supercrit Fluids. 2012; 72:36–45.
Salehi A, Fallah S, Zitterl-Eglseer K, Kaul HP, Surki AA, Mehdi B. Effect of organic fertilizers on antioxidant activityand bioactive compounds of fenugreek seeds in intercropped systems with buckwheat. Agronomy. 2019; 9(7):367.

Published:
License
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
